Scroll to:
Identification of key miRNAs as regulatory biomarkers of gonadotropins leading to infertility in males
https://doi.org/10.17749/2313-7347/ob.gyn.rep.2023.398
Abstract
Introduction. Infertility is a highly fatal reproductive system disorder that affects the ability of a couple to reproduce. Over the past decades, a drastic uplift has been recorded in infertility cases among males ranging from 20 to 70 % indicating spermatogenesis impairment.
Aim: to identify key microRNAs (miRNAs) as regulatory biomarkers of gonadotropins involved in dysregulation of fertility-related genes to propose potential therapeutic strategies that would combat the action of oncogenic miRNAs (oncomiRs).
Materials and Methods. Interaction analysis was performed between miRNAs and fertility-related genes namely luteinizing hormone choriogonadotropin receptor (LHCGR), gonadotropin-releasing hormone receptor (GnRHR), follicle-stimulating hormone receptor (FSHR) and cystic fibrosis transmembrane conductance regulator (CFTR) to identify key miRNAs as regulatory biomarkers of gonadotropins leading to infertility in males.
Results. A total of 10, 13, 31 and 18 strong and potential binding sites were predicted for miRNAs-LHCGR, miRNAs-GnRHR, miRNAs-FSHR, and miRNAs-CFTR respectively employing miRWalk (comprehensive genetic database including miRNA targets) followed by identification of 6, 18, 55 and 17 significant interactions through RNA22. Subsequently shortlisted miRNAs and messenger RNA (mRNA) regions were subjected to Vfold-Pipeline and RNAComposer individually for 3D structure prediction. Additionally molecular docking was carried out between miRNAs and mRNAs models that discovered potential and stable interactions elucidating miR-6880-FSHR(R2) as a highly stable complex with least binding affinity (-566.3) and high confidence score (0.999).
Conclusion. Hence this study proposes key oncomiRs as a diagnostic biomarker and therapeutic target to bring about a promising treatment strategy against male factor infertility. However wet lab investigations are required for further validations of proposed study.
Keywords
For citations:
Oohayyed N., Mohammed M., Al-Rahim A., Al Chalabi R., Shaban S., Suleiman A. Identification of key miRNAs as regulatory biomarkers of gonadotropins leading to infertility in males. Obstetrics, Gynecology and Reproduction. 2023;17(5):607-624. https://doi.org/10.17749/2313-7347/ob.gyn.rep.2023.398
Introduction / Введение
Infertility in men (azoospermia) has been considered a major health concern being highly prevalent worldwide indicating abnormalities in sperm parameters. Male infertility refers to impaired reproductive health due to defective semen parameters and lower testosterone levels. According to World Health Organization statistics, about 10–15 % of couples have been reported with infertility however male infertility contributes to half (50.0 %) of the infertility cases [1]. Infertility poses a considerable effect on psychological health and financial condition of patients producing economic burden on health care systems. Moreover, a higher mortality rate was observed among men having infertility factor than fertile men indicating greater risk of cancer among infertile individuals [2].
Infertility is caused by several risk factors that are categorized as acquired, idiopathic and congenital [3]. Acquired factors comprise exogenous factors (chemotherapy, radiation, heat, medications), severe diseases (renal failure, liver cirrhosis), acquired hypogonadotropic hypogonadism and varicocele which is a most common acquired factor of infertility with incidence rate of 40 % [4]. Moreover idiopathic factors incorporate obesity, psychological stress, smoking cigarettes, excessive intake of drugs, advanced paternal age, dietary practices and environmental exposure to toxins. Whereas, congenital factors (genetic factors) include chromosomal abnormalities, resulting in impaired testicular function and microdeletions in Y chromosome leading to spermatogenic abnormalities [5].
About 15 to 30 % of male infertility has been reported as a consequence of genetic factors including Y chromosome microdeletions and single gene defects [6]. Y chromosome is a male determining chromosome since most of its genes are specific to male fertility such as SRA (serum resistance associated) gene coupled with DAZ, RBMY, BDY (well known male fertility factors) regulate development of male genitalia. However microdeletions of fertility-related genes give rise to male-factor infertility anomalies [7]. Whereas single gene disorders occur as a result of mutations in a single gene for instance cystic fibrosis disorder (lung disease) is caused by recessive mutation in cystic fibrosis transmembrane conductance regulator (CFTR) that is responsible for congenital absence of the vas deferens (if inherited from both parents) leading to male infertility [8].
Hormones are the signaling molecules that regulate physiology and behavior of mammals sending signals into the bloodstream and tissues. The hypothalamic-pituitary-gonadal (HPG) axis (male reproductive hormone axis) is involved in regulation of male reproductive system through three major components namely hypothalamus, pituitary gland and gonads [9]. Furthermore, the gonadotropin-releasing hormone (GnRH) released by the hypothalamus binds to the gonadotropin-releasing hormone receptor (GnRHR), which is expressed on the gonadotrope cell surface that produces gonadotropins – follicle-stimulating hormone (FSH) and luteinizing hormone (LH) [10]. Gonadotropins are glycoprotein hormones involved in the regulation of the reproductive system and production of gametes and sex steroids hence essential for normal growth, sexual development and reproduction. However abnormal functioning of gonadotropins causes hypogonadism that occurs when sex glands do not produce enough hormones for normal functioning of the reproductive system [11]. Additionally malfunctioning hypothalamus and pituitary glands produce hypogonado-tropic hypogonadism. It refers to the condition in which the hypothalamus and pituitary gland do not produce hormones including FSH, LH and GnRH which are essential for the stimulation of sex glands [12]. Therefore, it is a prerequisite to identify the key microRNAs (miRNAs) involved in the downregulation of essential gonadotropins.
MicroRNAs (consisting of 18 to 22 nucleotides) are known as non-coding RNAs involved in the post transcriptional regulation of gene expression through either mRNA (matrix RNA) degradation or translational repression [13]. MicroRNAs have been emerged as disease biomarkers due to their influence on gene expression as tumor suppressors or oncogenic miRNAs (oncomiRs). Generally oncomiRs (upregulated miRNAs) show elevated expression in cancer whereas tumor suppressor miRNAs are underexpressed therefore regulation of miRNAs for therapeutic purposes is a concern of ongoing research. In breast cancer, high expression of oncomiRs majorly miR-532, miR-19b and miR-20b has been observed indicating underexpression of tumor suppressor genes considerably RERG, PTPRG and PTEN [14]. Moreover, overexpression of miR142-3p has been reported in testicular germ cell tumors suggesting that miR142-3p act as an oncomiR inducing the tumorigenesis through downregulation of key target PTPN23 (tumor suppressor gene) [15]. Another study has reported the causative role of oncomiRs in non-obstructive azoospermia elucidating that overexpression of miR-100 and let-7b downregulates the expression of estrogen-alpha (ERα) among infertile patients when compared with fertile men [16]. Considering the potential role of oncomiRs in male infertility, significant oncomiRs that are involved in the downregulation of fertility-regulating gonadotropins – GnRHR, FSHR, CFTR and luteinizing hormone choriogonadotropin receptor (LHCGR) needs to be investigated as disease biomarkers.
GnRHR is a receptor protein located on the cell surface of pituitary gonadotropes. It has been involved in the transduction of signals from GnRH (released by hypothalamus) to synthesize and stimulate the secretion of FSH and LH [17]. However FSHR is a follicle-stimulating hormone receptor mediated by G protein that activates downstream signal transduction pathways such as PI3K-AKT and ERK1/ERK2 leading to cellular response [18]. Whereas LHCGR is a G protein-coupled receptor for LH and chorionic gonadotropin and contributes to activation of signals that affect cell development and function [19]. CFTR is involved in regulation of ion channels by maintaining salt and water level on surfaces of the organ therefore such ion channels are essential for the proper functioning of organs such as lungs and pancreas [20]. Additionally mutated CFTR has been associated with male infertility suggesting mutations in the CFTR gene accounts for 78 % of infertility cases with congenital bilateral absence of the vas deferens [21].
Nevertheless, this study proposes key oncomiRs as regulatory biomarkers that may be involved in male infertility by dysregulating the essential fertility-related genes. These miRNAs may suggest potential therapeutic targets for combating the oncomiRs. Moreover, the crucial interactions predicted between miRNA-mRNA complexes and their strong and stable conformations might suggest their diagnostic and prognostic significance in bio-medical research. Additionally, the early diagnosis and prognosis will not only alleviate infertility determinants but will also ensure general health and well-being. Hence, infertility oncomiRs exerts potential role in acting as diagnostic, prognostic, and eventually therapeutic biomolecules for future perspectives.
Aim: to identify key miRNAs as regulatory biomarkers of gonadotropins involved in dysregulation of fertility-related genes to propose potential therapeutic strategies that would combat the action of oncomiRs.
Materials and Methods / Материалы и методы
Overview / Краткое описание
In this study, binding sites of miRNAs were predicted on fertility-regulating genes, namely LHCGR, GnRHR, FSHR and CFTR. The interactions between miRNAs and aforementioned genes were analyzed with respect to binding potential in order to identify the key miRNAs involved in downregulation of fertility-regulating genes (Fig. 1).

Figure 1. Overall workflow of miRNA-mRNA interaction analysis.
Note: BP – binding probability;
AU – Adenine Uracil content.
Рисунок 1. Общий процесс анализа взаимодействия микроРНК-мРНК.
Примечание: BС – вероятность связывания;
AU – содержание аденин-урацила.
Binding site prediction of miRNAs on fertility-regulating genes / Прогнозирование сайта связывания микроРНК на генах, регулирующих фертильность
The binding sites of miRNAs were predicted on fertility-related key genes LHCGR, GnRHR, FSHR and CFTR employing miRWalk (http://mirwalk.umm.uni-heidelberg.de). MiRWalk provides key insights of miRNA-target interactions based on machine learning predictions and experimental validations. It enables the user to predict miRNA-target interactions by either single gene or miRNA search option or through an advanced option called target mining [22]. Single gene search option was utilized to predict binding sites of miRNAs on fertility-regulating genes individually. Binding predictions were shortlisted on the basis of binding energy < –30 kcal/mol, binding probability = 1 and Adenine Uracil (AU) content < 0.6 [23]. Least binding energy represents highest stability while highest binding probability denotes strong binding plausibility. However AU content refers to AU rich elements of gene located at 30 nucleotides upstream and downstream of the predicted binding site. Additionally, AU rich elements are well known destabilizing elements, therefore least AU rich elements estimate highly stable structure [24].
Alignment analysis between potential miRNAs and fertility-related genes / Анализ соответствия потенциальных микроРНК генам, связанным с фертильностью
Predicted shortlisted microRNAs were aligned against fertility-related genes through RNA22 (https://cm.jefferson.edu/rna22/Interactive/) for the identification of significant binding interactions between miRNAs and their respective genes [25]. RNA22 is based on supervised machine learning method that employs combined approach to predict significant interaction i.e. pattern based approach (patterns of known miRNAs are used to predict putative target regions) and estimation of folding energy [26]. Shortlisted microRNAs were aligned against fertility-related genes (LHCGR, GNRHR, FSHR and CFTR) individually employing default parameters such as sensitivity = 63 %, specificity = 6 %, seed size = 7, minimum number of paired-up bases in heteroduplex = 12, maximum folding energy = –12 kcal/mol. For target gene sequence, NCBI (National Center for Biotechnology Information) Gene database (https://www.mirbase.org) was accessed to retrieve gene sequences, LHCGR (NM_000233.4), GnRHR (NM_000406.3), FSHR (NM_000145.4) and CFTR (NM_000492.4) [27]. The individual gene sequences were fed to RNA22 along with their corresponding shortlisted miRNA sequences retrieved through miRBase database (https://www.mirbase.org) [28]. Subsequently resultant interactions of miRNAs and genes were shortlisted on the basis of least p-value i.e. p-value < 0.05. Furthermore, gene regions depicting maximum and significant binding interactions with shortlisted miRNAs were extracted from genes and plotted in terms of binding sites and folding energy using the scatterplot function of seaborn (v0.11.2) library in python.
3D structure prediction of fertility-related genes / Трехмерное прогнозирование структуры генов, связанных с фертильностью
Modeling of fertility-related genes was carried out through RNAComposer (v1.0) (https://rnacomposer.cs.put.poznan.pl) [29]. RNAComposer uses sequence information and secondary structure topology of genes for their 3D structure prediction employing a dedicated database consisting of 3D RNA fragments obtained through RNA FRABASE. RNAComposer generates the model by comparing 3D RNA fragments with secondary structure topology of the queried gene. It offers two modes of gene modeling i.e. interactive mode (for single gene modeling at once) and batch mode (for modeling of 10 genes at a time) [28]. Interactive mode was utilized for modeling of fertility-related genes (parameters such as input sequence up to 500 nt, CONTRAfold as a secondary structure prediction method).
Modeling of shortlisted miRNAs / Моделирование окончательного набора микроРНК
Shortlisted microRNAs (with respect to maximum binding interactions with shortlisted regions of genes) were subjected to modeling through Vfold Pipeline (http://rna.physics.missouri.edu/vfoldPipeline/) [30]. Vfold Pipeline is a statistical approach of modeling that offers 2D and subsequently 3D structure prediction of miRNAs along RNA folding thermodynamics stabilities employing sequence information [30]. Shortlisted microRNA sequences (individually) were fed to Vfold Pipeline using default parameters. It generated a 3D structure file (pdb format) that was used for further analysis.
Molecular docking between shortlisted miRNA models and fertility-related gene models / Молекулярный докинг-анализ между моделями окончательного набора микроРНК и моделями генов, связанных с фертильностью
MicroRNA models were docked against gene models using HDOCK (v2021) (http://hdock.phys.hust.edu.cn/) [31]. It predicts the interactions between receptor and ligand molecule through a hybrid algorithm that offers template-free and template based docking. It takes either amino acid sequence or protein structures as an input and generates interaction results in the form interactive visualizations showing stable conformations with least binding affinity [32]. Individual 3D structures of short-listed microRNAs and genes were submitted to HDOCK as ligands and receptors respectively. Highly stable conformation (showing least binding affinity and highest confidence score) of miRNA-mRNA docked model was shortlisted out of 10 confirmations for each complex. Furthermore, shortlisted miRNA-mRNA docked complexes were visualized through PyMOL (v2.5.4) (https://pymol.org/2/). PyMOL is a molecular visualization system based on the python programming language that is employed for 3D visualizations of macromolecules including nucleic acids and proteins [33].
Results / Результаты
Prediction of miRNA interactions against fertility-related genes / Прогнозирование взаимодействий микроРНК с генами, связанными с фертильностью
The miRWalk database was employed to identify microRNAs and predict stable and strong interactions between predicted microRNAs and fertility-related genes, considerably LHCGR, GnRHR, FSHR and CFTR. It was observed that a total of 10, 13, 31 and 18 microRNAs showed strong and stable interactions (binding probability = 1, binding energy < –30 kcal/mol, AU content < 0.6) against aforementioned genes respectively. The interactions of predicted microRNAs against each fertility-related gene are provided in Tables 1–4 and scatterplot function of seaborn.
Table 1. Predicted interactions between luteinizing hormone
choriogonadotropin receptor (LHCGR) and miRNAs through miRWalk.
Таблица 1. Прогнозируемые взаимодействия между геном рецептора
лютеинизирующего гормона/хориогонадотропина (LHCGR) и микроРНК
с использованием базы данных miRWalk.
miRNA id Идентификатор микроРНК | miRNA sequence Последовательность микроРНК |
miR-339-3p | UGAGCGCCUCGACGACAGAGCCG |
miR-3147 | GGUUGGGCAGUGAGGAGGGUGUGA |
miR-3677-3p | CUCGUGGGCUCUGGCCACGGCC |
miR-3714 | GAAGGCAGCAGUGCUCCCCUGU |
miR-3135b | GGCUGGAGCGAGUGCAGUGGUG |
miR-4689 | UUGAGGAGACAUGGUGGGGGCC |
miR-4739 | AAGGGAGGAGGAGCGGAGGGGCCCU |
miR-4745-5p | UGAGUGGGGCUCCCGGGACGGCG |
miR-4758-5p | GUGAGUGGGAGCCGGUGGGGCUG |
miR-6777-5p | ACGGGGAGUCAGGCAGUGGUGGA |
Table 2. Predicted interactions between gonadotropin-releasing
hormone receptor (GnRHR) and miRNAs through miRWalk.
Таблица 2. Прогнозируемые взаимодействия между геном рецептора
гонадотропин-рилизинг гормона (GnRHR) и микроРНК
с использованием базы данных miRWalk.
miRNA id Идентификатор микроРНК | miRNA sequence Последовательность микроРНК |
miR-638 | AGGGAUCGCGGGCGGGUGGCGGCCU |
miR-1538 | CGGCCCGGGCUGCUGCUGUUCCU |
miR-3620-5p | GUGGGCUGGGCUGGGCUGGGCC |
miR-4640-5p | UGGGCCAGGGAGCAGCUGGUGGG |
miR-4685-5p | CCCAGGGCUUGGAGUGGGGCAAGGUU |
miR-4749-5p | UGCGGGGACAGGCCAGGGCAUC |
miR-5001-3p | UUCUGCCUCUGUCCAGGUCCUU |
miR-5787 | GGGCUGGGGCGCGGGGAGGU |
miR-6089 | GGAGGCCGGGGUGGGGCGGGGCGG |
miR-6741-5p | GUGGGUGCUGGUGGGAGCCGUG |
miR-6752-5p | GGGGGGUGUGGAGCCAGGGGGC |
miR-6857-5p | UUGGGGAUUGGGUCAGGCCAGU |
miR-8069 | GGAUGGUUGGGGGCGGUCGGCGU |
Table 3. Predicted interactions between follicle-stimulating
hormone receptor (FSHR) and miRNAs through miRWalk.
Таблица 3. Прогнозируемые взаимодействия между геном рецептора
фолликулостимулирующего гормона (FSHR) и микроРНК
с использованием базы данных miRWalk.
miRNA id Идентификатор микроРНК | Duplex Дуплекс |
miR-211-3p | GCAGGGACAGCAAAGGGGUGC |
miR-296-3p | GAGGGUUGGGUGGAGGCUCUCC |
miR-762 | GGGGCUGGGGCCGGGGCCGAGC |
miR-939-5p | UGGGGAGCUGAGGCUCUGGGGGUG |
miR-1233-5p | AGUGGGAGGCCAGGGCACGGCA |
miR-1207-5p | UGGCAGGGAGGCUGGGAGGGG |
miR-3192-5p | UCUGGGAGGUUGUAGCAGUGGAA |
miR-4463 | GAGACUGGGGUGGGGCC |
miR-4632-5p | GAGGGCAGCGUGGGUGUGGCGGA |
miR-4651 | CGGGGUGGGUGAGGUCGGGC |
miR-4656 | UGGGCUGAGGGCAGGAGGCCUGU |
miR-1343-5p | UGGGGAGCGGCCCCCGGGUGGG |
miR-4739 | AAGGGAGGAGGAGCGGAGGGGCCCU |
miR-2467-3p | AGCAGAGGCAGAGAGGCUCAGG |
miR-4524b-5p | AUAGCAGCAUAAGCCUGUCUC |
miR-6075 | ACGGCCCAGGCGGCAUUGGUG |
miR-6722-3p | UGCAGGGGUCGGGUGGGCCAGG |
miR-6848-5p | UGGGGGCUGGGAUGGGCCAUGGU |
miR-6858-5p | GUGAGGAGGGGCUGGCAGGGAC |
miR-6880-5p | UGGUGGAGGAAGAGGGCAGCUC |
miR-6885-5p | AGGGGGGCACUGCGCAAGCAAAGCC |
miR-7112-3p | UGCAUCACAGCCUUUGGCCCUAG |
miR-4433b-3p | CAGGAGUGGGGGGUGGGACGU |
miR-1909-3p | CGCAGGGGCCGGGUGCUCACCG |
miR-1914-3p | GGAGGGGUCCCGCACUGGGAGG |
miR-4433a-3p | ACAGGAGUGGGGGUGGGACAU |
miR-4640-5p | UGGGCCAGGGAGCAGCUGGUGGG |
miR-4758-5p | GUGAGUGGGAGCCGGUGGGGCUG |
miR-5787 | GGGCUGGGGCGCGGGGAGGU |
miR-6791-5p | CCCCUGGGGCUGGGCAGGCGGA |
miR-6847-5p | ACAGAGGACAGUGGAGUGUGAGC |
Table 4. Predicted interactions between cystic fibrosis transmembrane
conductance regulator (CFTR) and miRNAs through miRWalk.
Таблица 4. Прогнозируемые взаимодействия между геном регулятора
трансмембранной проводимости при муковисцидозе (CFTR) и микроРНК
с использованием базы данных miRWalk.
miRNA id Идентификатор микроРНК | Duplex Дуплекс |
miR-145-5p | GUCCAGUUUUCCCAGGAAUCCCU |
miR-370-3p | GCCUGCUGGGGUGGAACCUGGU |
miR-383-5p | AGAUCAGAAGGUGAUUGUGGCU |
miR-671-5p | AGGAAGCCCUGGAGGGGCUGGAG |
miR-1471 | GCCCGCGUGUGGAGCCAGGUGU |
miR-3619-3p | GGGACCAUCCUGCCUGCUGUGG |
miR-3620-5p | GUGGGCUGGGCUGGGCUGGGCC |
miR-3663-5p | GCUGGUCUGCGUGGUGCUCGG |
miR-1343-3p | CUCCUGGGGCCCGCACUCUCGC |
miR-4726-3p | ACCCAGGUUCCCUCUGGCCGCA |
miR-4769-5p | GGUGGGAUGGAGAGAAGGUAUGAG |
miR-5195-3p | AUCCAGUUCUCUGAGGGGGCU |
miR-6089 | GGAGGCCGGGGUGGGGCGGGGCGG |
miR-6749-5p | UCGGGCCUGGGGUUGGGGGAGC |
miR-6750-5p | CAGGGAACAGCUGGGUGAGCUGCU |
miR-6775-5p | UCGGGGCAUGGGGGAGGGAGGCUGG |
miR-6810-3p | UCCCCUGCUCCCUUGUUCCCCAG |
miR-6821-5p | GUGCGUGGUGGCUCGAGGCGGGG |
Alignment analysis between shortlisted miRNAs and fertility-related genes / Анализ соответствия окончательного набора микроРНК генам, связанным с фертильностью
Significant alignments between shortlisted miRNAs and fertility-related genes have been obtained through RNA22. A total of 6 significant alignments were observed between shortlisted miRNAs and shortlisted regions of LHCGR gene while RNA22 predicted 18 significant interactions between shortlisted miRNAs and their corresponding gene (GnRHR) regions. Additionally miRNA-FSHR alignment analysis revealed that 55 microRNAs were significantly bound with FSHR how-ever 17 miRNA-CFTR interactions were obtained through miRNA-CFTR alignment analysis. Alignments between miRNAs and fertility-related genes elucidated miRNA binding sites on their respective genes indicating low folding energy and high significance.
Shortlisting of miRNA-binding regions of fertility-related genes / Окончательный набор связывающих микроРНК областей генов, ассоциированных с фертильностью
MicroRNA-binding regions of fertility-related genes indicating maximum binding interactions with microRNAs were shortlisted through scatterplot function that gene-rated scatter plots for miRNAs-genes interactions (Fig. 2–5). It was observed that LHCGR, GnRHR, FSHR contained 2 such regions that showed maximum binding interactions with shortlisted miRNAs however, a total of 3 regions were identified for CFTR having maximum interactions with miRNAs (Table 5). For gene LHCGR, out of 6 interactions, 3 miRNAs (hsa-miR-3135b, hsa-miR-4739, hsa-miR-6777-5p) were shortlisted on the basis of maximum interactions (Table 6). It was observed that all of three shortlisted microRNAs (hsa-miR-3135b, hsa-miR-4739 and hsa-miR-6777-5p) were significantly bound with region 1 of gene LHCGR nevertheless 2 of aforementioned microRNAs also showed significant interactions (hsa-miR-3135b, hsa-miR-4739) with region 2 of LHCGR. For gene GnRHR, a total of 5 microRNAs (hsa-miR-4749-5p, hsa-miR-4685-5p, hsa-miR-6857-5p, hsa-miR-6752-5p, hsa-miR-8069) out of 18 interactions have been determined with maximum bindings (Table 7). All of the 5 shortlisted microRNAs showed significant interactions with region 1 of GnRHR however significant interactions of 2 aforementioned microRNAs (hsa-miR-3135b, hsa-miR-4739) have been also observed with region 2 of GnRHR. Moreover, scatterplot of miRNA-FSHR alignments revealed that 21 microRNAs out of 55 interactions have been significantly bound with FSHR indicating maximum interactions (Table 8). However, 3 (hsa-miR-762, hsa-miR-296-3p, hsa-miR-4651) out of 21 microRNAs were significantly bound with region 1 while all of 21 aforestated microRNAs have shown interaction with region 2 of FSHR indicating 3 common microRNAs between both regions. Furthermore, a total of 8 microRNAs were shortlisted for miRNA-CFTR alignments with respect to maximum interactions (Table 9). Out of 8 shortlisted microRNAs, 2 microRNAs (hsa-miR-145-5p, hsa-miR-4769-5p) were bound with region 1 while 3 unique microRNAs (hsa-miR-1343-3p, hsa-miR-6775-5p, hsa-miR-1471) including common hsa-miR-4769-5p of region 1 have been found to show significant interactions with region 2. Additionally, significant interactions of 3 unique microRNAs (hsa-miR-6821-5p, hsa-miR-3620-5p, hsa-miR-3619-3p) have been identified against region 3 including 1 common microRNA hsa-miR-6775-5p of region 2.
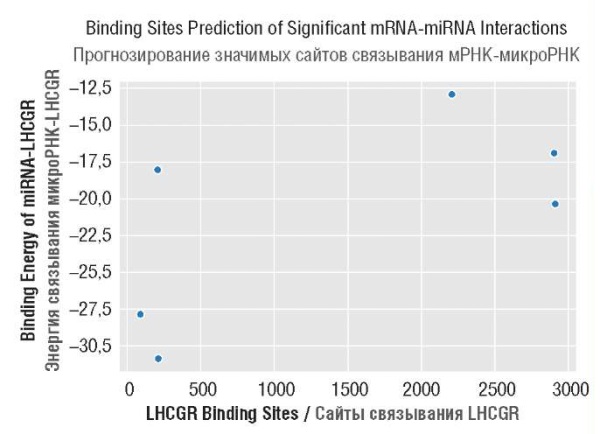
Figure 2. Scatter plot representation of significant miRNA-LHCGR interactions
for shortlisting of gene regions.
Note: LHCGR – luteinizing hormone choriogonadotropin receptor;
miRNA – microRNA.
Рисунок 2. Диаграмма рассеяния значимых взаимодействий микроРНК-LHCGR
для определения окончательного набора областей генов.
Примечание: LHCGR – рецептор лютеинизирующего
гормона/хориогонадотропина;
miRNA – микроРНК.

Figure 3. Scatter plot representation of significant miRNA-GnRHR interactions
for shortlisting of gene regions.
Note: GnRHR – gonadotropin-releasing hormone receptor;
miRNA – microRNA.
Рисунок 3. Диаграмма рассеяния значимых взаимодействий микроРНК-GnRHR
для определения окончательного набора областей генов.
Примечание: GnRHR – рецептор гонадотропин-рилизинг гормона;
miRNA – микроРНК.

Figure 4. Scatter plot representation of significant miRNA-FSHR interactions
for shortlisting of gene regions.
Note: FSHR – follicle-stimulating hormone receptor;
miRNA – microRNA.
Рисунок 4. Диаграмма рассеяния значимых взаимодействий микроРНК-FSHR
для определения окончательного набора областей генов
Примечание: FSHR – рецептор фолликулостимулирующего гормона;
miRNA – микроРНК.

Figure 5. Scatter plot representation of significant miRNA-CFTR interactions
for shortlisting of gene regions.
Note: CFTR – cystic fibrosis transmembrane conductance regulator;
miRNA – microRNA.
Рисунок 5. Диаграмма рассеяния значимых взаимодействий микроРНК-CFTR
для определения окончательного набора областей генов.
Примечание: CFTR – регулятор трансмембранной проводимости
при муковисцидозе;
miRNA – miRNA – микроРНК.
Table 5. Shortlisted regions of fertility-related genes.
Таблица 5. Окончательный набор областей генов,
связанных с фертильностью.
Gene / Ген | Region Область | Nucleotide length Длина, нуклеотиды |
LHCGR (luteinizing hormone choriogonadotropin receptor) LHCGR (рецептор лютеинизирующего гормона/хориогонадотропина) | Region 1 / Область 1 | 1–500 |
Region 2 / Область 2 | 2501–3000 | |
GnRHR (gonadotropin-releasing hormone receptor) GnRHR (рецептор гонадотропин-рилизинг-гормона) | Region 1 / Область 1 | 1–500 |
Region 2 / Область 2 | 501–1000 | |
FSHR (follicle-stimulating hormone receptor) FSHR (рецептор фолликулостимулирующего гормона) | Region 1 / Область 1 | 1–500 |
Region 2 / Область 2 | 1801–2300 | |
CFTR (cystic fibrosis transmembrane conductance regulator) CFTR (регулятор трансмембранной проводимости при муковисцидозе) | Region 1 / Область 1 | 1801-2300 |
Region 2 / Область 2 | 5001–5500 | |
Region 3 / Область 3 | 5501–6000 |
Table 6. Significant binding interactions between shortlisted miRNAs
and luteinizing hormone choriogonadotropin receptor (LHCGR) regions.
Таблица 6. Значимые связывающие взаимодействия
между окончательным набором микроРНК и областями рецептора
лютеинизирующего гормона/хориогонадотропина (LHCGR).
Gene region Область гена | miRNA микроРНК | Binding sites Сайты связывания | Folding energy Энергия связывания | р-value Значение р |
Region 1 Область 1 | miR-3135b | 88 | -27.8 | 0.0354 |
miR-4739 | 208 | -18.0 | 0.0318 | |
miR-6777-5p | 212 | -30.8 | 0.0318 | |
Region 2 Область 2 | miR-3135b | 2905 | -16.9 | 0.0215 |
miR-4739 | 2909 | -20.3 | 0.0215 |
Table 7. Significant binding interactions between shortlisted miRNAs
and gonadotropin-releasing hormone receptor (GnRHR) regions.
Таблица 7. Значимые связывающие взаимодействия
между окончательным набором микроРНК
и областями рецептора гонадотропин-рилизинг-гормона (GnRHR).
Gene region Область гена | miRNA микроРНК | Binding sites Сайты связывания | Folding energy Энергия связывания | р-value Значение р |
Region 1 Область 1 | miR-4749-5p | 19 | -15.9 | 0.0165 |
miR-4685-5p | 23 | -24.7 | 0.0165 | |
miR-6857-5p | 25 | -18.7 | 0.0165 | |
miR-6752-5p | 49 | -15.4 | 0.0165 | |
miR-8069 | 85 | -15.4 | 0.0165 | |
miR-6857-5p | 119 | -19.1 | 0.0165 | |
miR-4749-5p | 122 | -21.9 | 0.0165 | |
Region 2 Область 2 | miR-8069 | 697 | -22.8 | 0.0489 |
miR-6752-5p | 703 | -25.1 | 0.0489 |
Table 8. Significant binding interactions between shortlisted miRNAs
and follicle-stimulating hormone receptor (FSHR) regions.
Таблица 8. Значимые связывающие взаимодействия
между окончательным набором микроРНК
и областями рецептора фолликулостимулирующего гормона (FSHR).
Gene region Область гена | miRNA микроРНК | Binding sites Сайты связывания | Folding energy Энергия связывания | р-value Значение р |
Region 1 Область 1 | miR-762 | 253 | -15.2 | 0.0401 |
miR-296-3p | 254 | -15.6 | 0.0401 | |
miR-4651 | 254 | -18.5 | 0.0401 | |
Region 2 Область 2 | miR-4433b-3p | 1935 | -29.4 | 0.0123 |
miR-6880-5p | 1938 | -15.1 | 0.0123 | |
miR-6847-5p | 1938 | -14.4 | 0.0123 | |
miR-6722-3p | 1939 | -20.4 | 0.0123 | |
miR-1909-3p | 1939 | -14.7 | 0.0123 | |
miR-211-3p | 1940 | -16.54 | 0.0123 | |
miR-762 | 1946 | -20.5 | 0.0123 | |
miR-4739 | 1948 | -23.3 | 0.0123 | |
miR-1207-5p | 1949 | -20.3 | 0.0123 | |
miR-1914-3p | 1951 | -25.1 | 0.0123 | |
miR-1233-5p | 1953 | -22.6 | 0.0123 | |
miR-6848-5p | 1953 | -20.7 | 0.0123 | |
miR-296-3p | 1964 | -14.5 | 0.0123 | |
miR-1343-5p | 1965 | -15.0 | 0.0123 | |
miR-6858-5p | 1967 | -18.7 | 0.0123 | |
miR-4651 | 1999 | -15.8 | 0.0123 | |
miR-2467-3p | 1999 | -15.6 | 0.0123 | |
miR-4524b-5p | 2002 | -12.6 | 0.0123 | |
miR-4656 | 2064 | -15.5 | 0.0186 | |
miR-939-5p | 2251 | -18.7 | 0.0469 | |
miR-4758-5p | 2257 | -17.7 | 0.0469 |
Table 9. Significant binding interactions between shortlisted miRNAs
and cystic fibrosis transmembrane conductance regulator (CFTR) regions.
Таблица 9. Значимые связывающие взаимодействия
между окончательным набором микроРНК
и областями регулятора трансмембранной проводимости
при муковисцидозе (CFTR).
Gene region Область гена | miRNA микроРНК | Binding sites Сайты связывания | Folding energy Энергия связывания | р-value Значение р |
Region 1 Область 1 | miR-145-5p | 2087 | -14.1 | 0.00619 |
miR-4769-5p | 2162 | -12.3 | 0.00619 | |
miR-4769-5p | 2188 | -14.6 | 0.00619 | |
Region 2 Область 2 | miR-1343-3p | 5114 | -12.9 | 0.0431 |
miR-6775-5p | 5128 | -14.59 | 0.0431 | |
miR-4769-5p | 5160 | -12.4 | 0.0431 | |
miR-1471 | 5208 | -12.3 | 0.0431 | |
Region 3 Область 3 | miR-6821-5p | 5849 | -19.6 | 0.00749 |
miR-3620-5p | 5860 | -22.9 | 0.00749 | |
miR-6775-5p | 5904 | -22.9 | 0.00749 | |
miR-3619-3p | 5916 | -14.7 | 0.00749 |
Molecular docking analysis between 3D structures of shortlisted miRNAs and gene regions / Молекулярный докинг-анализ между трехмерными структурами окончательного набора микроРНК и областями генов
3D structures of shortlisted microRNAs and gene regions were modeled through Vfold Pipeline and RNAComposer respectively to perform molecular docking analysis. Subsequently predicted models of shortlisted miRNAs and fertility related genes were docked through Hdock-server for integrated protein–protein docking. Molecular docking generated 10 complexes for each miRNA-gene interaction; however only the top complex in terms of least docking score was shortlisted for further analysis. Out of 5, 7, 24 and 10 docked complexes for each interaction between shortlisted miRNAs and gene regions of LHCGR, GnRHR, FSHR and CFTR individually, only top docked complex was selected with respect to least affinity score (docking score) and highest confidence score indicating preferred and stable orientation of docked complex (Table 10). The docking results for each fertility-related gene were merged together and sorted on the basis of least docking score and highest confidence score in order to extract top 3 complexes out of all miRNAs-genes complexes. It was identified that miRNAs interactions with region 2 of FSHR majorly miR-6880-5p-R2, miR-2467-3-R2 and miR-211-3p-R2 were highly stable among all complexes with least docking score of -566.39, -554.35, -528.01 and highest confidence score of 0.9998, 0.9997 and 0.9995 respectively (Table 11).
Table 10. Shortlisted docked complexes
with respect to least docking score and highest confidence score.
Таблица 10. Окончательный набор пристыкованных комплексов
с учетом минимального показателя стыковки
и максимального показателя достоверности.
Gene / Ген | Complex Комплекс | Docking score Показатель стыковки | Confidence score Показатель достоверности |
LHCGR (luteinizing hormone choriogonadotropin receptor) LHCGR (рецептор лютеинизирующего гормона/хориогонадотропина) | miR-4739-R1 | -451.05 | 0.9976 |
GnRHR (gonadotropin-releasing hormone receptor) GnRHR (рецептор гонадотропин-рилизинг-гормона) | miR-6857-5p-R1 | -423.13 | 0.9958 |
FSHR (follicle-stimulating hormone receptor) FSHR (рецептор фолликулостимулирующего гормона) | miR-6880-5p-R2 | -566.39 | 0.9998 |
CFTR (cystic fibrosis transmembrane conductance regulator) CFTR (регулятор трансмембранной проводимости при муковисцидозе) | miR-4769-5p-R2 | -494.32 | 0.999 |
Table 11. Shortlisted top 3 docked complexes from merged docking results.
Таблица 11. Три лучших окончательных набора стыковочных комплексов
по объединенным результатам стыковки.
Gene / Ген | Complex Комплекс | Docking score Показатель стыковки | Confidence score Показатель достоверности |
FSHR (follicle-stimulating hormone receptor) FSHR (рецептор фолликулостимулирующего гормона) | miR-6880-5p-R2 | -566.39 | 0.9998 |
miR-2467-3p-R2 | -554.35 | 0.9997 | |
miR-211-3p-R2 | -528.01 | 0.9995 |
Visualization of shortlisted complexes / Визуализация окончательного набора комплексов
The shortlisted miRNA-mRNA complexes namely miR-4739-R1, miR-6857-5p-R1, miR-6880-5p-R2, miR-4769-5p-R2 were visualized through PyMOL to identify the interaction sites between shortlisted miRNAs and gene regions. A total of 20 interactions were observed between miR-4739 and region 1 of LHCGR with least binding affinity of -451.05 (Fig. 6) while 23 interactions were identified between miR-6857-5p and region 1 of GnRHR indicating least binding score of -423.13 (Fig. 7). However visualization analysis revealed that miR-6880-5p interacted with region 2 of FSHR at 20 different binding sites with least binding score i.e. -566.39 (Fig. 8) whereas 46 interactions were identified between miR-4769-5p and CFTR docked complex representing highest stability with least docking score of -494.32 (Fig. 9). Additionally visualization analysis of shortlisted complexes from merged docking analysis revealed highly stable complexes including aforementioned docked complex miR-6880-5p-R2 of FSHR (Fig. 8) and other 2 highly stable complexes namely miR-2467-3p-R2 and miR-211-3p-R2 of FSHR elucidating 46 and 22 stable interactions respectively (Fig. 10, 11).

Figure 6. Surface representation of docked complex – miR-4739-R1
of luteinizing hormone choriogonadotropin receptor (LHCGR)
indicating stable interactions between miR-4739 and region 1 of LHCGR:
(a) surface representation of docked complex;
(b) miRNA is shown in light pink color while mRNA
is represented in light purple however miRNA binding sites
and mRNA binding sites are indicated
in hot pink and dark blue colors respectively;
(c) polar contacts between ligand (miRNA) and receptor (mRNA)
are shown in yellow dotted lines
[drawn by the authors].
Рисунок 6. Изображение поверхности
пристыкованного комплекса – miR-4739-R1 рецептора
лютеинизирующего гормона/хориогонадотропина (LHCGR) указывает
на стабильные взаимодействия между miR-4739 и областью 1 LHCGR:
(а) изображение поверхности пристыкованного комплекса;
(b) микроРНК показана светло-розовым цветом,
мРНК – светло-фиолетовым цветом;
сайты связывания микроРНК и сайты связывания мРНК
обозначены ярко-розовым и темно-синим цветом соответственно;
(с) полярные контакты между лигандом (микроРНК) и рецептором (мРНК)
показаны желтыми пунктирными линиями
[рисунок авторов].

Figure 7. Surface representation of docked complex – miR-6857-R1
of gonadotropin-releasing hormone receptor (GnRHR)
indicating stable interactions between miR-6857 and region 1 of GnRHR:
(a) surface representation of docked complex;
(b) miRNA is shown in light pink color while mRNA
is represented in light purple however miRNA binding sites
and mRNA binding sites are indicated
in hot pink and dark blue colors respectively;
(c) polar contacts between ligand (miRNA) and receptor (mRNA)
are shown in yellow dotted lines
[drawn by the authors].
Рисунок 7. Изображение поверхности
пристыкованного комплекса – miR-6857-R1
рецептора гонадотропин-рилизинг гормона (GnRHR) указывает
на стабильные взаимодействия между miR-6857 и областью 1 GnRHR:
(а) изображение поверхности пристыкованного комплекса;
(b) микроРНК показана светло-розовым цветом,
мРНК – светло-фиолетовым цветом;
сайты связывания микроРНК и сайты связывания мРНК
обозначены ярко-розовым и темно-синим цветом соответственно;
(с) полярные контакты между лигандом (микроРНК) и рецептором (мРНК)
показаны желтыми пунктирными линиями
[рисунок авторов].

Figure 8. Surface representation of docked complex – miR-6880-R2
of follicle-stimulating hormone receptor (FSHR)
indicating stable interactions between miR-6880 and region 2 of FSHR:
(a) surface representation of docked complex;
(b) miRNA is shown in light pink color while mRNA
is represented in light purple however miRNA binding sites
and mRNA binding sites are indicated
in hot pink and dark blue colors respectively;
(c) polar contacts between ligand (miRNA) and receptor (mRNA)
are shown in yellow dotted lines
[drawn by the authors].
Рисунок 8. Изображение поверхности
пристыкованного комплекса – miR-6880-R2
рецептора фолликулостимулирующего гормона (FSHR) указывает
на стабильные взаимодействия между miR-6880 и областью 2 FSHR:
(а) изображение поверхности пристыкованного комплекса;
(b) микроРНК показана светло-розовым цветом цвет,
мРНК – светло-фиолетовым цветом;
сайты связывания микроРНК и сайты связывания мРНК
обозначены ярко-розовым и темно-синим цветом соответственно;
(с) полярные контакты между лигандом (микроРНК) и рецептором (мРНК)
показаны желтыми пунктирными линиями
[рисунок авторов].

Figure 9. Surface representation of docked complex – miR-4769-R2
of cystic fibrosis transmembrane conductance regulator (CFTR)
indicating stable interactions between miR-4769 and region 2 of CFTR:
(a) surface representation of docked complex;
(b) miRNA is shown in light pink color while mRNA
is represented in light purple however miRNA binding sites
and mRNA binding sites are indicated
in hot pink and dark blue colors respectively;
(c) polar contacts between ligand (miRNA) and receptor (mRNA)
are shown in yellow dotted lines
[drawn by the authors].
Рисунок 9. Изображение поверхности
пристыкованного комплекса – miR-4769-R2
регулятора трансмембранной проводимости при муковисцидозе (CFTR)
указывает на стабильные взаимодействия между miR-4769 и областью 2 CFTR:
(а) изображение поверхности пристыкованного комплекса;
(b) микроРНК показана светло-розовым цветом,
мРНК – светло-фиолетовым цветом;
сайты связывания микроРНК и сайты связывания мРНК
обозначены ярко-розовым и темно-синим цветом соответственно;
(с) полярные контакты между лигандом (микроРНК) и рецептором (мРНК)
показаны желтыми пунктирными линиями
[рисунок авторов].

Figure 10. Surface representation of docked complex – miR-2467-R2
of follicle-stimulating hormone receptor (FSHR)
indicating stable interactions between miR-2467 and region 2 of FSHR:
(a) surface representation of docked complex;
(b) miRNA is shown in light pink color while mRNA
is represented in light purple however miRNA binding sites
and mRNA binding sites are indicated
in hot pink and dark blue colors respectively;
(c) polar contacts between ligand (miRNA) and receptor (mRNA)
are shown in yellow dotted lines
[drawn by the authors].
Рисунок 10. Изображение поверхности
пристыкованного комплекса – miR-2467-R2 рецептора
фолликулостимулирующего гормона (FSHR) указывает
на стабильные взаимодействия между miR-2467 и областью 2 FSHR:
(а) изображение поверхности пристыкованного комплекса;
(b) микроРНК показана светло-розовым цветом,
мРНК – светло-фиолетовым цветом;
сайты связывания микроРНК и сайты связывания мРНК
обозначены ярко-розовым и темно-синим цветом соответственно;
(с) полярные контакты между лигандом (микроРНК) и рецептором (мРНК)
показаны желтыми пунктирными линиями
[рисунок авторов].

Figure 11. Surface representation of docked complex – miR-211-R2
of follicle-stimulating hormone receptor (FSHR)
indicating stable interactions between miR-211 and region 2 of FSHR:
(a) surface representation of docked complex;
(b) miRNA is shown in light pink color while mRNA
is represented in light purple however miRNA binding sites
and mRNA binding sites are indicated
in hot pink and dark blue colors respectively;
(c) polar contacts between ligand (miRNA) and receptor (mRNA)
are shown in yellow dotted lines
[drawn by the authors].
Рисунок 11. Изображение поверхности
пристыкованного комплекса – miR-211-R2 рецептора
фолликулостимулирующего гормона (FSHR) указывает
на стабильные взаимодействия между miR-211 и областью 2 FSHR:
(а) изображение поверхности пристыкованного комплекса;
(b) микроРНК показана светло-розовым цветом,
мРНК – светло-фиолетовым цветом;
сайты связывания микроРНК и сайты связывания мРНК
обозначены ярко-розовым и темно-синим цветом соответственно;
(с) полярные контакты между лигандом (микроРНК) и рецептором (мРНК)
показаны желтыми пунктирными линиями
[рисунок авторов].
Discussion / Обсуждение
Infertility is a highly prevalent global health problem affecting the reproductivity of 8–12 % of the couples having unprotected intercourse [34]. However, a global survey has reported a rapid increase in male infertility cases over the past decades ranging from 20 to 70 % [35]. It has been identified that males with reported infertility ailments are more prone to other diseases including metabolic disorders and several cancers affecting the overall wellbeing of the patients and posing financial pressure on patients and healthcare systems [2]. Infertility arises in men as a consequence of several risk factors: considerably acquired factors (radiations, severe disorders, hypogonadism and varicocele), idiopathic actors (chemical exposure, smoking, over-age, psychological problems) and congenital factors (genetic factors) [3]. Among various risk factors, genetic factors majorly Y chromosome microdeletions and single gene defects contribute largely to male infertility with a high incidence rate of 15 to 30 % [6]. Gonadotropins (fertility-regulating genes) namely LHCGR, GnRHR, FSHR in addition to wild CFTR have been reported to be responsible for normal regulation of the reproductive system. However dysregulation, particularly underexpression of aforestated genes may lead to infertility causing abnormalities in sperm parameters and reduced level of testosterone [11]. OncomiRs (upregulated miRNAs) are known hallmarks of male infertility causing downregulation of essential gonadotropins and have been investigated as disease biomarkers [14]. Considering the role of dysregulated gonadotropins as a consequence of interacting oncomiRs, miRNA-mRNA interactions are required to be identified and analyzed.
The miRNA-mRNA interaction analysis identified potential oncomiRs indicating strong interactions with fertility-related genes considerably LHCGR, GnRHR, FSHR and CFTR. Among miRNA-mRNA interactions, 10 strong interactions were observed for miRNAs-LHCGR, 13 against miRNAs-GnRHR, 31 for miRNAs-FSHR and 18 for miRNAs-CFTR indicating the binding potential of shortlisted microRNAs on aforesaid genes. Additionally binding sites were identified, elucidating that mutations in the binding sites may alter its target specificity and its subsequent impact on gene regulation [36]. However significant interactions between shortlisted miRNAs and fertility-related genes were identified using RNA22 tool that depicted 6, 18, 55 and 17 significant interactions for miRNAs-LHCGR, miRNAs-GnRHR, miRNAs-FSHR and miRNAs-CFTR respectively. RNA22 selects miRNAs on the basis of folding energy and p-value representing significant interactions. It has been reported that the most important factor for the significant miRNA-mRNA interaction is having the perfect homology between the seed sequence of miRNA and the corresponding target gene sequence, that can be acquired through RNA22 [37].
For LHCGR, interacting miRNAs majorly miR-3135b, miR-4739, miR-6777-5p were found to be associated with male infertility and related cancers. Dysregulated expression of predicted miR-3135b has been observed in pancreatic ductal adenocarcinoma causing tumor development and progression [38]. Several studies have confirmed that male infertility has been linked with the emergence of various malignancies majorly breast cancer, pancreatic cancer and testicular germ cell cancer [39][40]. Thus predicted miR-3135b can be used as a promising target for early diagnosis and prognosis of male infertility. While miR-4739 has been associated with spermatogenic impairment in the male Korean population elucidating its potential interaction with stromal antigen 3 (STAG3), however mutations in STAG3 effect its protein function leading to male infertility due to meiotic arrest [41]. It was observed that dysregulation of miR-6777 negatively modulates LINC00355:8 that in turn activates the Wnt/β-catenin signaling pathway and induces epithelial-mesenchymal transition (EMT) progression leading to hepatocellular carcinoma [42]. However hypogonadism is commonly found in patients having chronic liver disease thus considering the involvement of liver malignancies in inducing male infertility, miR-6777-5p may be considered a diagnostic biomarker and therapeutic target.
For miRNAs-GnRHR interactions, male infertility-related microRNAs have been identified including miR-4749, miR-4685-5p and miR-8069. The oncogenic function of miR-4749 was observed in impairment of tumor suppressor gene p53 that affects DNA binding function bringing about tumor progression and invasiveness [43]. Moreover oncogenic role of another predicted oncomiR miR-4685-5p has been identified in early stage lung adenocarcinoma [44] that reveals that miR-4685-5p may act as a potential signature for early diagnosis and prognosis of male infertility having correlation with CFTR mutations in lung cancer that modulate the risk of male infertility [45]. Elevated level of miR-8069 has been observed in pancreatic cancer that shows association with male infertility suggesting its oncogenic role [46].
For miRNAs-FSHR interactions, miR-762 has been known for its crucial role in mammalian reproduction. It interacts with 3 untranslated region (UTR) of DNA-damage related gene called RNF4 and alters its expression inducing growth and proliferation of porcine immature Sertoli cells inhibits apoptosis by activating DNA damage repair and minimizing the expression of androgen receptor [47]. Another study has confirmed the oncogenic role of miR-762 in non-obstructive azoospermia (NOA) as compared to fertile men, in which overexpression of miR-762 has been associated with NOA disease [48]. The underexpression of predicted miR-296 has been reported in highly fertile men suggesting that overexpression of miR-296 leads to infertility [49]. Additionally, elevated level of miR-296 expression has been also predicted in patients diagnosed with several classes of male factor infertility particularly teratozoospermia, asthenozoospermia, oligozoospermia and normozoospermia indicating characteristic role of miR-296 in inducing infertility [50]. Among impaired spermatogenesis cases of male Korean population, miR-4739-STAG3 interaction has been observed however STAG3 gene is a known hallmark of male factor infertility therefore predicted miR-4739 can be used as therapeutic target against male infertility [41].
For miRNAs-CFTR interactions, a study investigated the function of miR-145 by predicting its interacting genes in patients with spermatogenesis. It was observed that overexpression of miR-145 downregulates expression of male sex-determining gene SOX9 that in turn induces male fertility. Therefore the oncogenic role of miR-145 reveals its diagnostics and prognostic value as an infertility bio-marker [51]. miR-1471 was found to be overexpressed in patients with sertoli-cell-only syndrome that is characterized by azoospermia therefore miR-1471 can be used as a promising strategy against male infertility [52]. Overexpression of miR-1343 has been identified in capacitated spermatozoa through expression profiling study of capacitated spermatozoa and non-capacitated spermatozoa [53]. However, capacitated spermatozoa refers to essential physiological changes that are required for spermatozoa to undergo therefore miR-1343 can be used as a therapeutic target for mitigating the effect of infertility. Hence predicted microRNAs have shown significant association with male infertility and can be used as therapeutic targets for early diagnosis and prognosis of male infertility.
The mRNA regions of fertility-related genes were shortlisted with respect to maximum binding interactions with miRNAs that resulted in 2 regions for LHCGR, GnRHR, FSHR and 3 regions for CFTR. It was observed that LHCGR, GnRHR, FSHR and CFTR showed maximum binding interactions with 3 miRNAs, 5 miRNAs, 21 miRNAs and 8 miRNAs respectively. 3D structures of shortlisted microRNAs and mRNA regions were docked through HDOCK to determine the best conformation of the docked complex in terms of high stability. Molecular docking generated 5 complexes for miRNAs-LHCGR, 7 complexes for miRNAs-GnRHR, 24 complexes for miRNAs-FSHR and 10 complexes for miRNAs-CFTR. However, only the top complex with respect to highest confidence score and least affinity score was considered for further analysis. All of the shortlisted complexes namely miR-4739-LHCGR(R1), miR-6857-5p-GnRHR(R1), miR-6880-FSHR(R2) and miR-4769-5p-CFTR(R2) revealed interaction of miRNAs with coding sequence (CDS) region of respective genes. Moreover top 3 complexes particularly miR-6880-FSHR(R2), miR-2467-3p-FSHR(R2), miR-211-3p-FSHR(R2) were shortlisted from combined docking results containing all shortlisted complexes making up a total of 46 complexes. Among shortlisted complexes, miRNA interaction was observed at the CDS region of corresponding genes. It has been reported that binding regions particularly 3 UTR, 5 UTR and CDS have functional impact on regulatory activity of microRNAs for instance miRNA interaction with 3 UTR region of gene causes mRNA degradation while miRNA binding with 5 UTR or CDS region of genes is responsible for translational repression through mRNA silencing [54]. Therefore, mRNA binding sites are required to be identified in order to predict functional characteristics of miRNAs in male infertility.
In this study binding affinities of shortlisted microRNAs were identified against fertility-related genes majorly LHCGR, GnRHR, FSHR and CFTR. Out of all individual and combined complexes, miR-6880-FSHR(R2) was found to be a highly stable complex indicating least binding affinity (-566.3) and high confidence score (0.999); however miR-4769-5p-CFTR(R2) exhibited high number of interactions at 46 different sites comparatively with docking score of -494.32 and confidence score of 0.999. Hence strong and potential interactions between miRNAs and mRNAs have determined the stability and potential efficacy of aforementioned complexes.
Conclusively this research has predicted highly significant and stable interactions between oncomiRs and fertility-regulating genes thus unveil the causative determinants of azoospermia. Additionally microRNA-binding regions of genes indicating significantly high interactions with oncomiRs were identified that determines dysregulated expression of gonadotropins. Regulatory role of predicted microRNAs has been investigated in several malignancies including pancreatic ductal adenocarcinoma, hepatocellular carcinoma, lung adenocarcinoma, teratozoospermia, asthenozoospermia, oligozoospermia, normozoospermia and spermatogenic impairment. Additionally characteristic involvement of gonadotropins in fertility has been reported in the regulation of fertility in males therefore suppressing the role of oncomiRs can alter the spermatogenesis impairment that improves the health of patients. Hence this study proposes a promising strategy to identify disease biomarkers that would mitigate infertility issues in men. However wet lab investigations are required to validate the findings of the proposed study.
Conclusion / Заключение
In this study miRNA-mRNA interaction analysis has revealed significant interactions of 6, 18, 55 and 17 between miRNAs and genes particularly LHCGR, GnRHR, FSHR and CFTR respectively. Subsequently molecular docking between shortlisted mRNA regions and miRNA generated stable complexes however only top one highly stable conformation and top 3 best poses were shortlisted from individual and combined complexes respectively that disclosed miR-6880-FSHR(R2) as a highly stable complex indicating least binding affinity (-566.3) and high confidence score (0.999). Additionally shortlisted complexes revealed that miRNAs interacted with genes at CDS region indicating translational repression. Thus strong interactions between miRNAs and mRNAs have ensured the stability and potential efficacy of aforementioned complexes. Furthermore oncogenic role of predicted miRNAs has been discovered in fertility-related disorders majorly teratozoospermia, asthenozoospermia, oligozoospermia, normozoospermia and spermatogenic impairment. Therefore it has been inferred from the study that predicted miRNAs may act as oncomiRs suppressing the function of gonadotropins. Hence this study proposes key oncomiRs that can be used as potential candidates for infertility diagnosis and prognosis.
References
1. Babakhanzadeh E., Nazari M., Ghasemifar S., Khodadadian A. Some of the factors involved in male infertility: a prospective review. Int J Gen Med. 2020;13:29–41. https://doi.org/10.2147/IJGM.S241099.
2. Hanson B.M., Eisenberg M.L., Hotaling J.M. Male infertility: a biomarker of individual and familial cancer risk. Fertil Steril. 2018;109(1):6–19. https://doi.org/10.1016/j.fertnstert.2017.11.005.
3. Okonofua F.E., Ntoimo L.F.C., Omonkhua A. et al. Causes and risk factors for male infertility: a scoping review of published studies. Int J Gen Med. 2022;15:5985–97. https://doi.org/10.2147/IJGM.S363959.
4. White W.M., Mobley J.D., Kim E.D. Varicocele: Practice Essentials, History of the Procedure, Problem. Medscape, 2023. Available at: https://emedicine.medscape.com/article/438591-overview.
5. Carson S.A., Kallen A.N. Diagnosis and management of infertility. JAMA. 2021;326(1):65–76. https://doi.org/10.1001/jama.2021.4788.
6. Sudhakar D.V.S., Shah R., Gajbhiye R.K. Genetics of male infertility – present and future: A narrative review. J Hum Reprod Sci. 2021;14(3):217–27. https://doi.org/10.4103/jhrs.jhrs_115_21.
7. Colaco S., Modi D. Genetics of the human Y chromosome and its association with male infertility. Reprod Biol Endocrinol. 2018;16(1):14. https://doi.org/10.1186/s12958-018-0330-5.
8. Wong R., Gu K., Ko Y., Patel P. Congenital absence of the vas deferens: cystic fibrosis transmembrane regulatory gene mutations. Best Pract Res Clin Endocrinol Metab. 2020;34(6):101476. https://doi.org/10.1016/j.beem.2020.101476.
9. Silva M.S.B., Giacobini P. New insights into anti-Müllerian hormone role in the hypothalamic–pituitary–gonadal axis and neuroendocrine development. Cell Mol Life Sci. 2021;78(1):1–16. https://doi.org/10.1007/s00018-020-03576-x.
10. Kaiser U.B., Sabbagh E., Katzenellenbogen R.A. et al. A mechanism for the differential regulation of gonadotropin subunit gene expression by gonadotropin-releasing hormone. Proc Natl Acad Sci U S A. 1995;92(26):12280–4. https://doi.org/10.1073/pnas.92.26.12280.
11. Plunk E.C., Richards S.M. Endocrine-disrupting air pollutants and their effects on the hypothalamus-pituitary-gonadal axis. Int J Mol Sci. 2020;21(23):9191. https://doi.org/10.3390/ijms21239191.
12. Fink J., Schoenfeld B.J., Hackney A.C. et al. Human chorionic gonadotropin treatment: a viable option for management of secondary hypogonadism and male infertility. Expert Rev Endocrinol Metab. 2021;16(1):1–8. https://doi.org/10.1080/17446651.2021.1863783.
13. Cangiano B., Swee D.S., Quinton R., Bonomi M. Genetics of congenital hypogonadotropic hypogonadism: peculiarities and phenotype of an oligogenic disease. Hum Genet. 2021:140:(1):77–111. https://doi.org/10.1007/s00439-020-02147-1.
14. Yao Q., Chen Y., Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol. 2019;51:11–7. https://doi.org/10.1016/j.cbpa.2019.01.024.
15. Di Palo A., Siniscalchi C., Salerno M. et al. What microRNAs could tell us about the human X chromosome. Cell Mol Life Sci. 2020;77(20):4069–80. https://doi.org/10.1007/s00018-020-03526-7.
16. Batool A., Liu X.-M., Zhang C.-L. et al. Recent advances in the regulation of testicular germ cell tumors by microRNAs. Front Biosci (Landmark Ed). 2019:24(4):765–76. https://doi.org/10.2741/4749.
17. Munawar M., Liaqat I., Ali S. et al. MicroRNAs and male infertility. In: Recent Advances in Noncoding RNAs. Ed. L. Tutar. IntechOpen, 2022. https://doi.org/10.5772/intechopen.106757. Available at: https://www.intechopen.com/chapters/83297.
18. Casteel C., Singh G. Physiology, gonadotropin-releasing hormone. StatPearls, 2022. Available at: https://www.ncbi.nlm.nih.gov/books/NBK558992/.
19. Haldar S., Agrawal H., Saha S. et al.Overview of follicle stimulating hormone and its receptors in reproduction and in stem cells and cancer stem cells. Int J Biol Sci. 2022;18(2):675–92. https://doi.org/10.7150/ ijbs.63721.
20. Mann O.N., Kong C.-S., Lucas E.S. et al. Expression and function of the luteinizing hormone choriogonadotropin receptor in human endometrial stromal cells. Sci Rep. 2022;12(1):8624. https://doi.org/10.1038/s41598-022-12495-9.
21. Hanssens L.S., Duchateau J., Casimir G.J. CFTR protein: not just a chloride channel? Cells. 2021;10(11):2844. https://doi.org/10.3390/ cells10112844.
22. Cioppi F., Rosta V., Krausz C. Genetics of azoospermia. Int J Mol Sci. 2021;22(6):3264. https://doi.org/10.3390/ijms22063264.
23. Sticht C., Torre C.D.L., Parveen A., Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018;13(10):e0206239. https://doi.org/10.1371/journal.pone.0206239.
24. Li D., Knox B., Gong B. et al. Identification of translational microRNA biomarker candidates for ketoconazole-induced liver injury using nextgeneration sequencing. Toxicol Sci. 2021;179(1):31–43. https://doi.org/10.1093/toxsci/kfaa162.
25. Barreau C., Paillard L., Osborne H.B. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33(22):7138–50. https://doi.org/10.1093/nar/gki1012.
26. Loher P., Rigoutsos I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics. 2012;28(24):3322–3. https://doi.org/10.1093/ bioinformatics/bts615.
27. Brown G.R., Hem V., Katz K.S. et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015;43(Database issie):D36–42. https://doi.org/10.1093/nar/gku1055.
28. Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. https://doi.org/10.1093/nar/gky1141.
29. Biesiada M., Purzycka K.J., Szachniuk M. et al. Automated RNA 3D structure prediction with RNAComposer. Methods Mol Biol. 2016;1490:199–215. https://doi.org/10.1007/978-1-4939-6433-8_13.
30. Li J., Zhang S., Zhang D., Chen S.-J. Vfold-Pipeline: a web server for RNA 3D structure prediction from sequences. Bioinformatics. 2022;38(16):4042–3. https://doi.org/10.1093/bioinformatics/btac426.
31. Yan Y., Zhang D., Zhou P.et al. HDOCK: A web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(W1):W365–W373. https://doi.org/10.1093/nar/gkx407.
32. Li H., Huang E., Zhang Y. et al. HDOCK update for modeling protein-RNA/ DNA complex structures. Protein Science. 2022;31(11):e4441. https://doi.org/10.1002/pro.4441.
33. Yuan S., Chan H.C.S., Hu Z. Using PyMOL as a platform for computational drug design. WIREs Comput Mol Sci. 2017;7(2):e1298. https://doi.org/10.1002/wcms.1298.
34. Agarwal A., Baskaran S., Parekh N. et al. Male infertility. Lancet. 2021;397(10271):319–33. https://doi.org/10.1016/S0140-6736(20)32667-2.
35. Agarwal A., Finelli R., Selvam M.K.P. et al. A global survey of reproductive specialists to determine the clinical utility of oxidative stress testing and antioxidant use in male infertility. World J Mens Health, 2021;39(3):470– 88. https://doi.org/10.5534/wjmh.210025.
36. Jafarinejad-Farsangi S., Jazi M.M., Rostamzadeh F., Hadizadeh M. High affinity of host human microRNAs to SARS-CoV-2 genome: an in silico analysis. Noncoding RNA Res. 2020;5(4):222–31. https://doi.org/10.1016/j.ncrna.2020.11.005.
37. Mukherjee M., Goswami S. Global cataloguing of variations in untranslated regions of viral genome and prediction of key host RNA binding proteinmicroRNA interactions modulating genome stability in SARS-CoV-2. PLoS One. 2020;15(8):e0237559. https://doi.org/10.1371/journal.pone.0237559.
38. Aita A., Millino C., Sperti C. et al. Serum miRNA profiling for early PDAC diagnosis and prognosis: a retrospective study. Biomedicines. 2021;9(7):845. https://doi.org/10.3390/biomedicines9070845.
39. Nagirnaja L., Aston K., Conrad D. The genetic intersection of male infertility and cancer. Fertil Steril. 2018;109(1):20–6. https://doi.org/10.1016/j.fertnstert.2017.10.028.
40. Swerdlow A.J., Bruce C., Cooke R. et al. Infertility and risk of breast cancer in men: a national case–control study in England and Wales. Breast Cancer Res. 2022;24(1):29. https://doi.org/10.1186/s13058-022-01517-z.
41. Nam Y., Kang K. M., Sung S.R. et al. The association of stromal antigen 3 (STAG3) sequence variations with spermatogenic impairment in the male Korean population. Asian J Androl. 2020;22(1):106–11. https://doi.org/10.4103/aja.aja_28_19.
42. Zhou F., Lei Y., Xu X. LINC00355:8 promotes cell proliferation and migration with invasion via the MiR-6777-3p/Wnt10b axis in Hepatocellular Carcinoma. J Cancer. 2020;11(19):5641–55. https://doi.org/10.7150/jca.43831.
43. Bizzarri A.R., Cannistraro S. Investigation of a direct interaction between miR4749 and the tumor suppressor p53 by fluorescence, FRET and molecular modeling. Biomolecules. 2020;10(2):346. https://doi.org/10.3390/biom10020346.
44. Chen Z., Wei J., Li M., Zhao Y. A circular RNAs dataset landscape reveals potential signatures for the detection and prognosis of early-stage lung adenocarcinoma. BMC Cancer. 2021:21(1):781. https://doi.org/10.1186/s12885-021-08293-7.
45. Kamiński P., Baszyński J., Jerzak I. et al. External and genetic conditions determining male infertility. Int J Mol Sci. 2020;21(15):5274. https://doi.org/10.3390/ijms21155274.
46. Yoshizawa N., Sugimoto K., Tameda M. et al. MiR-3940-5p/miR-8069 ratio in urine exosomes is a novel diagnostic biomarker for pancreatic ductal adenocarcinoma. Oncol Lett. 2020;19(4);2677–84. https://doi.org/10.3892/ol.2020.11357.
47. Reza A.M.M.T., Choi Y.-J., Han S.G. et al. Roles of microRNAs in mammalian reproduction: From the commitment of germ cells to periimplantation embryos. Biol Rev Camb Philos Soc. 2019;94(2);415–38. https://doi.org/10.1111/brv.12459.
48. Abu-Halima M., Hammadeh M., Schmitt J. et al. Altered microRNA expression profilesof human spermatozoa inpatients with different spermatogenic impairments. Fertil Steril. 2013;99(5):1249–55.e16. https://doi.org/10.1016/j.fertnstert.2012.11.054.
49. Alves M.B.R., Celeghini E.C.C., Belleannée C. From sperm motility to sperm-borne microRNA signatures: new approaches to predict male fertility potential. Front Cell Dev Biol. 2020;8:791. https://doi.org/10.3389/ fcell.2020.00791.
50. Tomic M., Bolha L., Pizem J. et al. Association between sperm morphology and altered sperm microRNA expression. Biology (Basel). 2022;11(11):1671. https://doi.org/10.3390/biology11111671.
51. Zhang L., Ding X., Nie S. et al.Association of hsa-miR-145 overexpression in human testicular cells with male infertility. Mol Med Rep. 2015;11(6):4365–72. https://doi.org/10.3892/mmr.2015.3273.
52. Gunes S., Arslan M.A., Hekim G.N.T., Asci R. The role of epigenetics in idiopathic male infertility. J Assist Reprod Genet. 2016;33(5):553–69. https://doi.org/10.1007/s10815-016-0682-8.
53. Sahoo B., Choudhary R.K., Sharma P. et al. Significance and relevance of spermatozoal RNAs to male fertility in livestock. Front Genet. 2021;12:768196. https://doi.org/10.3389/fgene.2021.768196.
54. Wang J., Liu S., Shi J. et al. The role of miRNA in the diagnosis, prognosis, and treatment of osteosarcoma. Cancer Biother Radiopharm. 2019;34(10):605–13. https://doi.org/10.1089/cbr.2019.2939.
About the Authors
N. A. OohayyedIraq
Noor A. Oohayyed – Assistant Lecturer, Department of Medical Biotechnology, College of Biotechnology
M. M. Mohammed
Iraq
Mais M. Mohammed – Assistant Lecturer
Scopus Author ID: 57561562000
A. M. Al-Rahim
Iraq
Aya M. Al-Rahim – Assistant Lecturer, Department of Medical Biotechnology, College of Biotechnology
Scopus Author ID: 57226461520
Researcher ID: ACZ-6702-2022
R. N. Al Chalabi
Iraq
Rawaa N. Al Chalabi – Assistant Professor, Dr, Department of Medical Biotechnology, College of Biotechnology
Scopus Author ID: 57210340073
Researcher ID: ABD-7380-2020
S. A. Shaban
Russian Federation
Semaa A. Shaban – PhD, Assistant Professor, Department of Biology, College of Sciences
Scopus Author ID: 57223083825
Researcher ID: H-1150-2019
A. A. J. Suleiman
Iraq
Ahmed A. J. Suleiman – PhD in Biotechnology and Molecular Biology, Professor, Department of Biotechnology, College of Science
Scopus Author ID: 57200943508
Researcher ID: AAF-7727-2019
Review
For citations:
Oohayyed N., Mohammed M., Al-Rahim A., Al Chalabi R., Shaban S., Suleiman A. Identification of key miRNAs as regulatory biomarkers of gonadotropins leading to infertility in males. Obstetrics, Gynecology and Reproduction. 2023;17(5):607-624. https://doi.org/10.17749/2313-7347/ob.gyn.rep.2023.398

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.











































